Graphs have been successfully used to model structured and unstructured data, such as social networks, citation networks, and biological networks, and analyze their characteristics w.r.t. various metrics and criteria. Computing these metrics and finding meaningful patterns inside large datasets has always been a fundamental problem, and several algorithms have been proposed for this purpose. Yet, some of these existing algorithms are not suitable for today's data, since it is dynamic, frequently updated, and often have temporal properties. To this end, we analyze, engineer, develop, and improve existing or novel scalable algorithms on large graphs for various mining and analysis purposes including community detection, link prediction, and ranking/popularity measures.
Related Publications

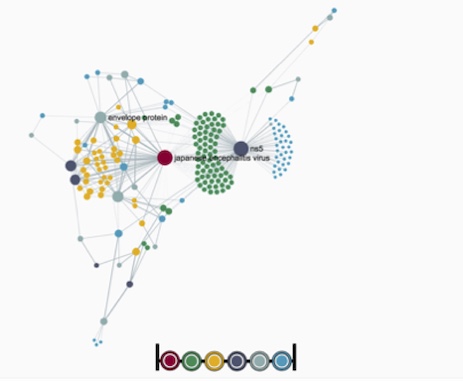

In collaboration with Dr. Janies’s Lab, at UNCC, we are developing novel techniques for analysis and visualization of spreads of emergent pathogens. My group develops graph mining, network analysis techniques and high performance computing solutions to scale those computations. We develop workflows for phylogeographic analysis and visualization of the pathogens of interest that provide information on the geographic traffic of the spread of the pathogens and places that are hubs for their transport.
Related PublicationsResearchers typically rely on manual methods to discover new research such as keyword-based search on search engines, which suffers from the confusion induced by different names of identical concepts in different fields. We developed a method, direction-aware random walk with restart, to suggest relevant papers using the relations in citation graph. Using the metadata harvested mainly from open-access databases, we implemented a scalable web interface of the service (accessible via http://theadvisor.osu.edu). In addition, we observed that the nonzero pattern of the citation matrix is highly irregular and the computation suffers from high number of cache misses. We propose techniques for storing the matrix in memory efficiently and reducing the number of cache misses.
Related Publications



With the advent of next-generation high throughput sequencing instruments, large volumes of short sequence data are generated at an unprecedented rate. Processing and analyzing these massive data requires overcoming several challenges including mapping of generated short sequences to a reference genome. Our research in this area focuses on parallelization of short sequence mapping process. We also develop analytical models to explain run time costs of various parallelization schemes in order to understand the trade-offs among them.
Related PublicationsBiological network analysis is interested in utilizing heterogeneous biological data, such as transcriptomics, proteomics, and gene-disease relationship datasets. Our research mainly focuses on the problem of data integration, summarization, and extraction of meaningful patterns from such molecular networks. It involves building unstructured, high-level representations of the interactions between biological agents and formulating and validating hypotheses on the intrinsic properties of this data.
Related Publications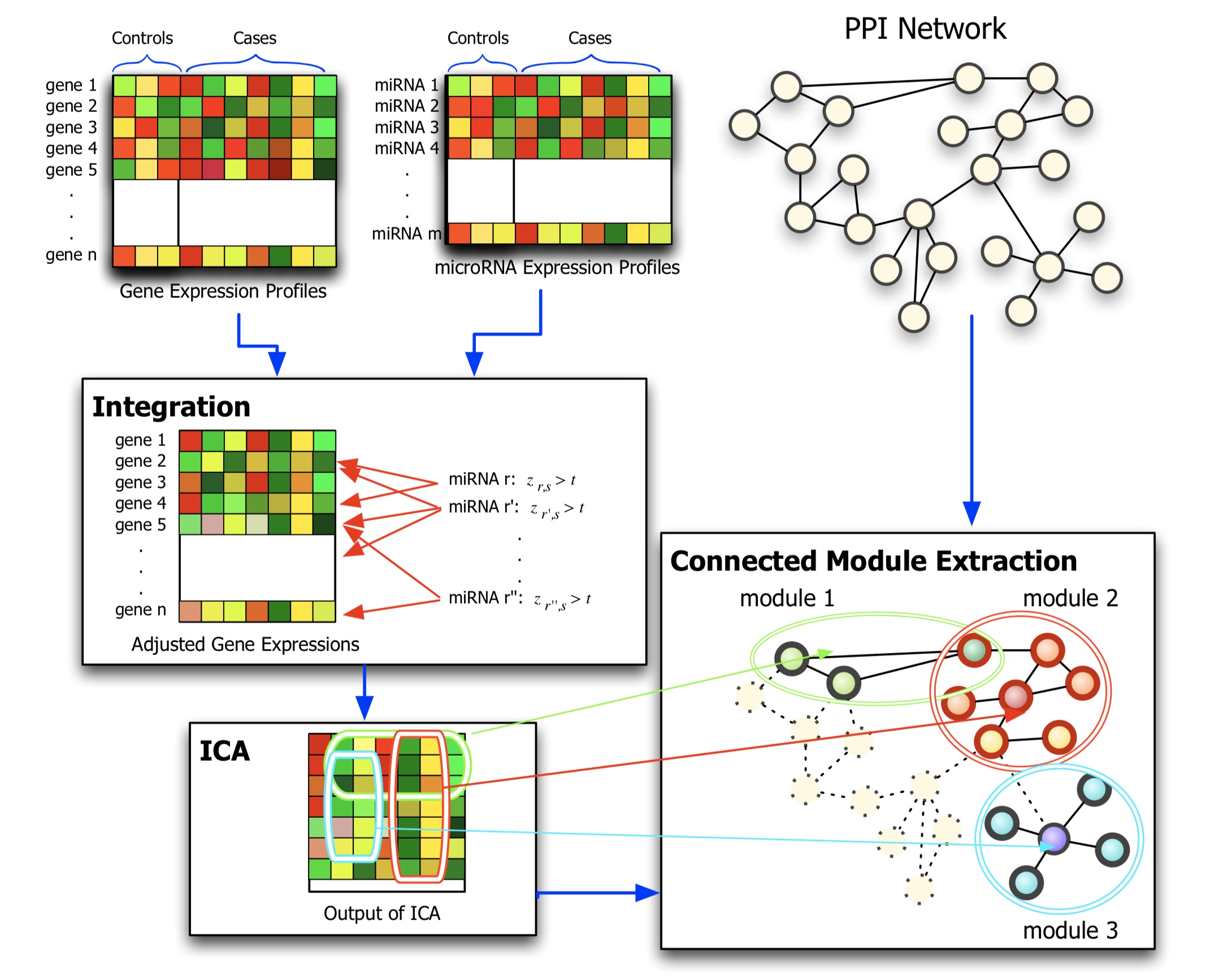

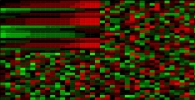

Biclustering is a very popular method to detect hidden co-regulation patterns among genes. At the HPC Lab, we develop novel biclustering algorithms to identify subsets of genes with high correlation by reducing false negatives due to spurious or unrelated samples in microarray datasets. One of the major applications of our biclustering methods is to reveal genes and proteins important in the complex process of breast tumor formation.
Related PublicationsOur work in this area is targeted at easing the use of complex multicore accelerator architectures, such as GPUs and the Cell processor. High-performance scientific applications naturally require a large amount of processing power, and modern cluster supercomputers are increasingly being built with accelerator co-processors. Our work helps to bridge the gap between traditional HPC programming methods and accelerators by using component-based programming frameworks and new runtime technology. Recent work includes DataCutter-Lite, a light-weight component-based middleware for multi-to-many core architectures, such as IBM Cell Broadband Engine and multi-core CPUs.
Related Publications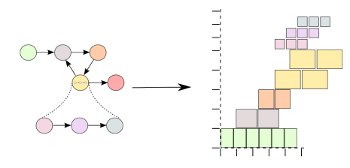

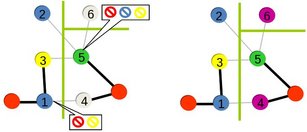

One of our recent projects is development of distributed memory graph coloring algorithms. In many parallel scientific computing applications coloring is used to identify independent tasks that can be performed concurrently. In such cases graph is typically distributed across processors and hence the coloring needs to be computed in parallel. We have started with distance-1 and distance-2 coloring algorithms, and currently we are investigating how we can scale this algorithms to machines with tens of thousands of processors.
Related Publications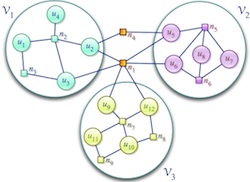

Today, many large scientific computations are carried out on large distributed memory parallel computers. One of the first steps is to compute a distribution of computation and data across the processors in a manner that achieves high parallel efficiency. This problem is known as the (static) load-balancing (a.k.a partitioning). In parallel adaptive applications, the computational structure of the applications changes over time, leading to load imbalances even though the initial load distributions were balanced. To restore balance and to keep communication volume low in further iterations of the applications, dynamic load-balancing (repartitioning) of the changed computational structure is required. In collaboration with Zoltan Team of Sandia National Laboratories, we develop parallel hypergraph partitioning-based methods and tools for these load-balancing problems.
Related PublicationsParallel sparse matrix-vector multiplication is one of the most important kernels in parallel iterative methods including solvers for linear systems, linear programs, eigensystems, and least squares problems. During the last decade, we have proposed several, successful hypergraph-based models and methods for communication-reducing partitioning of sparse matrices for parallel matrix vector multiplication. Currently we are working on developing scalable parallel implementations of these approaches as well as easy-to-use high-level interfaces for application developers, such as Matlab interface.
A typical first step of a direct solver for linear system Mx=b is reordering of symmetric matrix M to improve execution time and space requirements of the solution process. We are working on developing novel nested-dissection-based ordering approaches that utilizes hypergraph partitioning.
Related Publications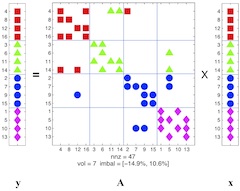

In many distributed applications, the computations take place in a discrete 2D or 3D space. At each iteration of the application, the state of each cell is updated using the state of neighbor cells in a stencil fashion. In applications such as particle-in-cell simulation and raycasting, the computation time required to update each cell may vary in function of well known parameters (such as the distribution of the particle in the field or the position of the objects in the scene). In this project, we are tackling the distribution of such applications on a parallel computer. The cells must be allocated to the processor meticulously by taking into account both the load balance between the processors and the communication time between two adjacent cells. Methods should also take into account the possible dynamic aspects of such systems. To solve this problem, we are proposing new rectangular decomposition algorithms and analyze their behavior both theoretically and practically.
Related Publications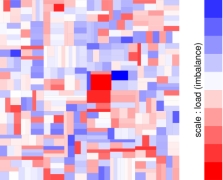

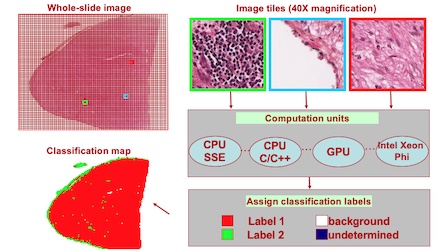

Our goal is to develop a computer-aided diagnosis and prognosis systems that will assist the pathologists in the grading of various cancer types.
Related PublicationsDirected Acyclic Graph (DAG) scheduling is a very important problem in distributed memory computing. There are three main metrics that determine the performance of a scheduling algorithm: (1) runtime complexity, (2) length of generated schedules and (3) number of processors used by the generated schedules. It is extremely difficult to simultaneously minimize these three metrics, hence there are trade-offs offered by various algorithms. In our lab, we develop new DAG scheduling algorithms by carefully accounting for mentioned trade-offs to best address the needs of the applications being considered.
Related Publications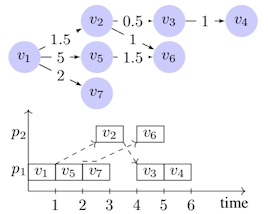

In many fields of science, engineering, and medicine, data is being collected and generated at an increasing rate, thanks mainly to high-resolution measurements made possible by advanced sensor technologies and large scale simulations enabled by inexpensive, high-performance computing through commodity PC clusters. Hence, scientific research is increasingly becoming data driven. In most of the cases, data is stored in shared data resources and, usually parts of it, needs to be transferred to compute resource for analysis. We develop locality-aware scheduling algorithms for data-intensive tasks, and efficient data transfer mechanisms for coordinated data-transfer over wide-area networks.
Related PublicationsThe goals of this project are (1) to explore in- novative nuclear physics algorithms that address the scale explosion in nuclear problems and hence, to make solutions feasible; (2) to exploit highly efficient computational methods for applications to nuclear physics and other fields in physics; (3) to perform early experiments with petascale archi- tectures; and, (4) to solve ultra-large realistic problems at a level of accuracy needed to test the underlying strong interaction theory together with its associated symmetries and to supply nuclear structure information key to modeling astrophysical processes.
Related PublicationsIn many fields of science, engineering, and medicine, research is increasingly becoming more and more data-intensive. Simulation and computation have become new tools of scientific discovery. Scientists are able to run simulations that almost match the complexity of the real world. In addition to the computational power of the systems, advances in sensor technologies have enabled capturing of high-throughput data, such as gene micro-arrays. Storage capacity has shown an even faster trend than processing power, continuing almost to triple every eighteen months. Hence, vast amounts of data are being generated and collected at an increasing rate. Development of applications that are able to process and analyze these data, however, is becoming extremely difficult due both to the ever-growing complexity of the datasets and the complications involved in new, high-performance computing platforms. We design and develop middleware tools that will make it easier to store, retrieve and analyze large, distributed datasets.
Related PublicationsInitially motivated by the computational challenges in histopathological image analysis, we started to investigate use of emerging architectures such as Graphics Processing Units (GPUs) and IBM Cell Broadband Engine for accelerating the computations in other image analysis applications.
Related PublicationsIn addition to Parallel Graph Coloring and Parallel Hypergraph Partitioning, we have also investigated the parallelization of other graph kernels.
Related PublicationsOther Biomedical Informatics publications.
Related Publications